NTU-MIT research team speeds up drug development with AI
Published in Nature Nanotechnology, the breakthrough could cut vaccine and drug delivery timelines from months to weeks.
An AI model co-developed by Nanyang Technological University’s College of Computing and Data Science (CCDS) and the Massachusetts Institute of Technology (MIT) could transform how RNA-based medicines, including mRNA vaccines, are designed and delivered.
Published today in Nature Nanotechnology, the study introduces COMET, an AI “experiment simulator” that predicts the most effective formulations for lipid nanoparticles (LNPs); the microscopic “vehicles” that carry mRNA into cells. By identifying optimal designs in silico, COMET could reduce the need for extensive laboratory testing, cutting development timelines from months to weeks.
The model found nanoparticle designs that outperformed existing clinical benchmarks, signalling a major leap in RNA medicine delivery.

"This breakthrough showcases how AI can help make the development of new medicine faster, cheaper, and better,” said Assistant Professor Alvin Chan, who began the project as a postdoctoral researcher at MIT before continuing it at NTU.
By drastically reducing the real-world experiments needed, we can get effective medicines to patients faster, and at lower cost.
Solving the LNP challenge
If mRNA are the passengers, then lipid nanoparticles are the vehicles that deliver them safely to their destination inside our cells. Designing the best “vehicle” is a complex challenge — it’s not just about the type of material (lipids) used, but also the precise combination of components, their ratios, and how they’re assembled.
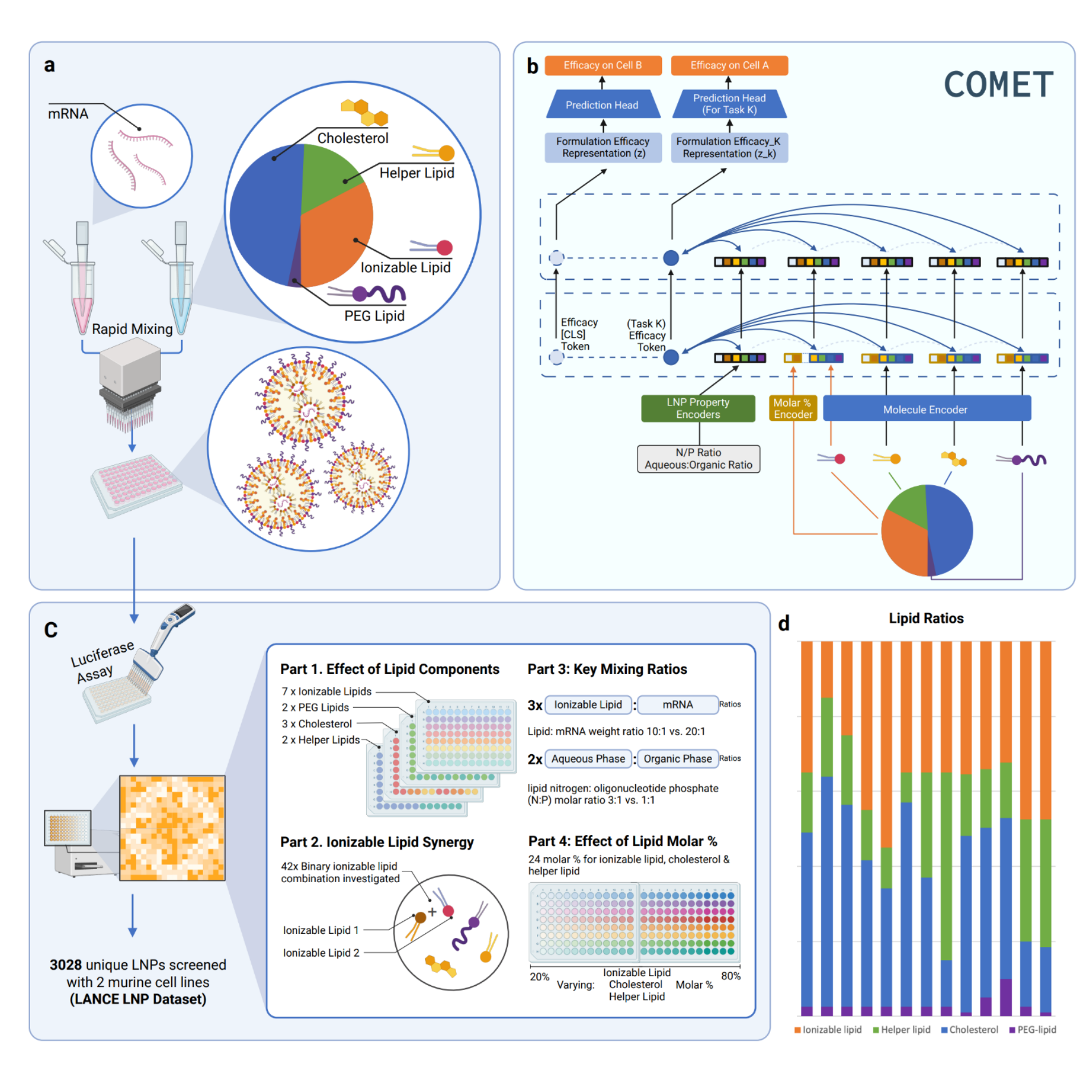
How COMET changes the game
Instead of making and testing thousands of physical samples, scientists can feed potential LNP “recipes” into COMET, which predicts their efficacy for different applications. This makes it possible to identify the “needle in a haystack” — the best formulation among millions or even billions of possibilities — in a fraction of the time.
“This is where COMET becomes a general‑purpose technology for RNA medicines — whether you’re targeting different cells or improving stability, the same system can guide you to the best solution,” Chan explained.
The research combined AI with high‑throughput screening, creating an exceptionally rich dataset for COMET to train on and enabling it to explore a vast design space virtually. In validation tests, several COMET‑designed LNPs outperformed clinically approved benchmarks in both laboratory and animal studies.
From his postdoctoral years at MIT and Harvard Medical School to his current role at NTU’s College of Computing and Data Science, Asst Prof Alvin Chan has built teams on both sides of the globe united by a shared goal: harnessing AI to transform drug discovery.
.png?sfvrsn=fcc807de_1)
.png?sfvrsn=197aa068_1)
Beyond pandemics
While pandemic preparedness is a clear application, COMET’s potential extends far beyond infectious diseases. Its predictive power could help scientists develop RNA therapies that target specific organs or cell types, opening new possibilities for treating cancers and other complex diseases.
What’s Next
Chan’s team is now developing an AI‑powered platform that designs the most effective lipid nanoparticle for any chosen therapeutic target, paving the way for RNA medicines that can reach and treat diseases in specific organs or cell types with greater precision and safety.
About the study
- COMET is trained on one of the largest and most diverse datasets of lipid nanoparticle (LNP) formulations ever compiled, capturing variations in materials, ingredient ratios, and manufacturing methods.
- Built on a transformer‑based neural network architecture (similar to advanced language models), COMET can detect complex relationships between nanoparticle design and delivery performance.
- In validation tests, several COMET‑designed LNPs outperformed clinically approved benchmarks in both laboratory and animal studies.
- The model is application‑flexible — able to adapt designs for different organs or cell types — making it a potential general‑purpose platform for RNA‑based medicines.
- Alvin’s team is also developing high‑throughput experimental technologies that can test hundreds of RNA formulations in a single animal, accelerating the feedback loop between AI design and real‑world validation.
You may read the full paper here.




7c137751-1960-4051-9719-3e607d92ca71.tmb-listing.jpg?Culture=en&sfvrsn=9cfc5323_1)

