Chromatin Biochemistry and Biophysics

DESCRIPTION OF LAB RESEARCH WORK
Mammalian telomeres consist of the tandemly arranged DNA sequence repeats TTAGGG that act as a platform for the recruitment of the capping structure at the end of the linear chromosomes, which protect their ends from incorrect DNA repair and degradation. In humans, this consists of the six-protein Shelterin complex as well as nucleosomes that package 5–15 kbp of telomeric DNA into chromatin. Aberrant regulation of telomeres has been linked with cancer development and ageing-associated pathologies. However, almost nothing was known about the structure of telomeric chromatin at the molecular level. Recently (Soman et al, Nature, 699, 1048, 2022), we characterized telomeric chromatin fibres with EM and single-molecule magnetic tweezers. We determined the Cryo-EM structure of the condensed telomeric tetra-nucleosome chromatin fibre, which revealed a novel columnar architecture.
A general aim of our research is the unravelling of the principles and control of the packaging of DNA in the cell nucleus within the context of chromatin with emphasis to the unique telomeric chromatin. We are interested in understanding the molecular interactions and the (electrostatic) mechanisms that induce, stabilize and regulate chromatin folding and what are the implications of this for telomere maintenance and its relation to cancer and aging.
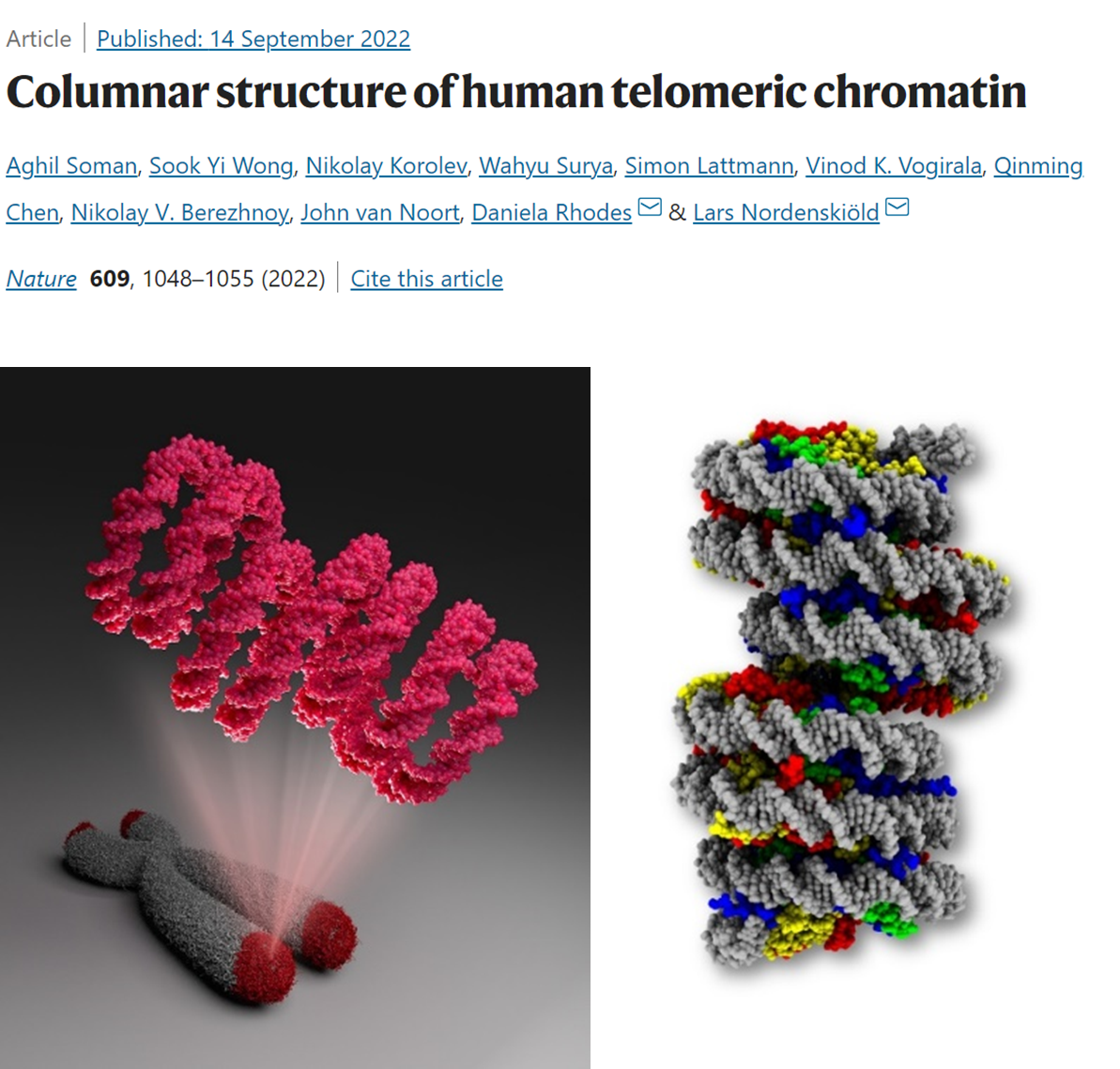
Fig. 1.
We solved the mystery of chromatin organization at telomeres (left), revealing a columnar packaging of nucleosomes (right). The structure is also the first higher-order chromatin structure formed on a native human DNA and diverges from textbook models (Soman et al, Nature, 699, 1048, 2022).
9ea80211-5e40-4ec9-98e7-b8492d8f8c8f.jpg?sfvrsn=770807ba_3)
Fig. 2.
DNA phase separation with formation of hexagonally ordered DNA liquid crystals (A and B) and DNA toroid (C and D.) Results from bottom-up multiscale computer simulations using the Inverse Monte Carlo method. (Sun et al, NAR, 47(11), 5550-5562, 2019)
A general aim of our research is the unravelling of the principles and control of the packaging of DNA in the cell nucleus within the context of chromatin with emphasis to the unique telomeric chromatin. We are interested in understanding the molecular interactions and the (electrostatic) mechanisms that induce, stabilize and regulate chromatin folding and what are the implications of this for gene regulation
LAB MEMBERS
 | LEAD PI
Lars Nordenskiöld Chair, School of Biological Sciences Professor, School of Biological Sciences Email: larsnor@ntu.edu.sg Phone: (65) 6592 7506 Office:SBS-01N-01 |
 | Nikolay Korolev
Principal Research Fellow Email: korolev@ntu.edu.sg |
 | Sun Tiedong
Senior Research Fellow Email: tdsun@staff.main.ntu.edu.sg |
 | Aghil Soman
Research Fellow Email: asoman@ntu.edu.sg |
 | Wayne Shum Research Associate Email: WShum@ntu.edu.sg |
CURRENT PROJECTS
Some specific aims are:
- To experimentally and theoretically investigate the folding of recombinant in vitro prepared nucleosome core particles (NCP) and well-defined chromatin in the form of nucleosome arrays and how this is affected by changes of the histone tail modifications and transcription factor binding with applications to telomeric chromatin.
- To identify the difference in structure and dynamics between telomeric and “normal” canonical nucleosomes and chromatin, using biophysical and structural methods like X-ray crystallography, EM and solid-state NMR (Fig. 1).
- To characterise the binding and structural and dynamic properties of HP1 interaction with telomeric chromatin and compare it with canonical chromatin
- To establish the physical basis in terms of electrostatic/molecular interactions for DNA and chromatin phase separations (LLPS), it’s structure and dynamics using computer modelling tools (Fig. 2).
These aims are realized by using a combination of molecular biology, biochemical and chemical synthesis techniques in combination with biophysical approaches such as single molecule measurements (magnetic tweezers), solid-state NMR, X-ray structure determination, synchrotron x-ray diffraction (SAXS), analytical ultracentrifugation, as well as well as Cry-Electron Microscopy (EM) methods. Experimental work is often performed in combination with our computer modelling studies.
Subprojects:
- Telomeric chromatin: Unravelling the structural properties of the telomeric nucleosome and chromatin and how it differs from canonical chromatin.
- Understanding the structural and dynamic properties of condensed heterochromatin within the context of telomeres. Specifically, the role of heterochromatin protein 1 (HP1) by solid-state NMR study of heterochromatin maintenance by HP1 protein at telomeres
- Multi-scale computer modeling of electrostatic interactions in DNA, the nucleosome core particle (NCP), chromatin up to mesoscale megabase size chromosomal chromatin for an understanding of chromatin liquid liquid phase separation (LLPS).
PUBLICATIONS
- Xiangyan Shi, Bhuvaneswari Kannaian, Chinmayi Prasanna, Aghil Soman, Lars Nordenskiöld, (2023), Solid-state NMR Study of the Structure and Dynamics of Histone H2B in a Nucleosome Core Particle, Accepted, Communications Biology, 6(1), 672, 10.1038/s42003-023-05050-3
- Aghil Soman, Sook Yi Wong, Nikolay Korolev, Wahyu Surya, Simon Lattmann, Vinod K. Vogirala, Qinming Chen, Nikolay V. Berezhnoy, John van Noort, Daniela Rhodes, and Lars Nordenskiöld, Columnar structure of human telomeric chromatin, Nature, 609, 1048–1055, 2022
- Korolev, N., Zinchenko. A., Soman, A., Chen, Q., Wong, S. Y., Berezhnoy, N. V., Basak, R., Van Der Maarel, J. R. C., van Noort, J., Nordenskiöld, L.. (2022). “Reconstituted TAD-size chromatin fibers feature heterogeneous nucleosome clusters”. Scientific Reports, 12:15558 (10.1038/s41598-022-19471-3)
- Tiedong Sun, Vishal Minhas, Alexander Mirzoev, Nikolay Korolev, Alexander P. Lyubartsev and Lars Nordenskiöld, (2022) “A Bottom-Up Coarse-Grained Model for Nucleosome-Nucleosome Interactions with Explicit Ions”, Journal of Chemical Theory and Computation (ACS), 18, 3948−3960
- Qinming Chen, Lei Zhao, Aghil Soman, Anastasia Arkhipova, Jindi Li, Hao Li, Yinglu Chen, Xiangyan Shi and Lars Nordenskiöld, Chromatin liquid-liquid phase separation (LLPS) is regulated by ionic conditions and chromatin fibre length, Cells, 11, 3145, 2022
- Aghil Soman, Nikolay Korolev and Lars Nordenskiöld, Telomeric chromatin structure, Current Opinion in Structural Biology, 77:102492, 2022
- Aghil Soman, Chong Wai Liew, Hsiang Ling Teo, Nikolay Berezhnoy, Vincent Olieric, Nikolay Korolev, Daniela Rhodes, Lars Nordenskiöld, The Human Telomeric Nucleosome Displays Distinct Structural and Dynamic Properties, Nucleic Acids Research, 48(10), 5383–5396, 2020
- Tiedong Sun, Alexander Mirzoev, Vishal Minhas, Nikolay Korolev, Alexander P. Lyubartsev and Lars Nordenskiöld, “A multiscale analysis of DNA phase separation: from atomistic to mesoscale level”, Nucleic Acids Research, 47(11), 5550-5562, 2019
- Xiangyan Shi, Chinmayi Prasanna, Toshio Nagashima, Toshio Yamazaki, Konstantin Pervushin and Lars Nordenskiöld, Structure and Dynamics in the Nucleosome Revealed by Solid-State NMR, Angewandte Chemie, Int. Ed., 57,9734 –9738, 2018
- Nikolay Korolev, Alexander P. Lyubartsev and Lars Nordenskiöld “A systematic analysis of nucleosome core particle and nucleosome-nucleosome stacking structure”, Scientific Reports, 8:1543, |DOI:10.1038/s41598-018-19875-0, 2018
- Anatoly Zinchenko, Nikolay Berezhnoy, William Rosencrans, Nikolay Korolev, Johan RC van der Maarel and Lars Nordenskiöld, “Single-Molecule Compaction of Megabase-Long Chromatin Molecules by Multivalent Cations”, Nucleic Acids Research, 46(2) 635–649, 2018
- Qinming Chen, Renliang Yang, Nikolay Korolev, Chuan Fa Liu and Lars Nordenskiöld, Regulation of Nucleosome Stacking and Chromatin Compaction by the Histone H4 N-Terminal Tail - H2A Acidic Patch Interaction, Journal of Molecular Biology, 429(13), 2075-2092, 2017
- Abdollah Allahverdi, Yang Renliang, Nikolay Korolev, Yanping Fan, Curt Davey, Chuan Fa Liu and Lars Nordenskiöld, “The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association”, Nucleic Acids Research, 39(5): 1680-1691, 2011

